The largest synthetic genome has been born so far: not all codons are constructed
The largest synthetic genome has been born so far: not all codons are constructed
May 30, 2019 Source: Nature Natural Science Research
Window._bd_share_config={ "common":{ "bdSnsKey":{ },"bdText":"","bdMini":"2","bdMiniList":false,"bdPic":"","bdStyle":" 0","bdSize":"16"},"share":{ }};with(document)0[(getElementsByTagName('head')[0]||body).appendChild(createElement('script')) .src='http://bdimg.share.baidu.com/static/api/js/share.js?v=89860593.js?cdnversion='+~(-new Date()/36e5)];The largest synthetic genome to date has been born, and this genome with only a few amino acid-encoded codons makes it possible to encode proteins containing unnatural amino acid residues in the future.
Over the past decade, as the cost of DNA chemical synthesis has decreased and the way DNA fragments are assembled has become more sophisticated, synthetic biology has entered a new phase of synthesizing the entire chromosome and genome. To date, researchers have been able to construct synthetic DNA with up to 1 million base pairs, such as a set of chromosomes from Saccharomyces cerevisiae and various synthetic genomes of Mycoplasma mycoides. Today, Fredens et al. published a paper in Nature that the team synthesized a 4 million base pair Escherichia coli genome. The study is a landmark event in the emerging field of synthetic genomics and the first application of synthetic genomics technology to this “hard work†of bacteria.

Source: pixabay
Synthetic genomics has given us a whole new way of understanding the rules of life, while also pushing synthetic biology toward the direction of genome design. The pioneer in this field is the researchers from the Craig Venter Institute in the United States. In order to determine the minimum number of genes required for independent living cells, they chose to use this method first. The genomic fragments were redesigned, and these fragments were chemically synthesized and finally assembled. With this technology, these researchers have successfully reduced the genome size of Mycoplasma mycoplasma by about 50%. If you switch to the genome editing tool, it will greatly increase the workload. This has been confirmed by previous E. coli experiments - the gene deletion method can only remove up to 15% of the genome.
Fredens and colleagues used the reduced E. coli genome as a synthetic genomic template, but there is another minimization in their minds—reduction of codons. The genetic code itself is redundant: a total of 64 codons (base triplets) encode 20 amino acids, as well as "start" and "termination" points that represent the initiation and termination of a protein coding sequence. This redundancy means that there are six codons encoding serine and three possible stop codons. The protein coding sequence of E. coli has 64 available codons. Fredens et al. designed, synthesized and assembled the E. coli genome with only 61 codons. Their approach was to terminate the two serine codons and one. Codons are replaced by synonymous codons - synonymous codons are codons that have different "spelling" but give the same instruction. Previous studies using the genome editing tool successfully synthesized an E. coli with 63 codons, but this only required replacing the stop codon of the TAG sequence (321 in the genome) with another stop codon. This time, the reduction to 61 codons requires 18214 codons to be changed. For this reason, a combination of genes must be used.
Fredens and colleagues synthesized the E. coli genome through a large-scale DNA assembly and genome integration method previously developed to study the E. coli codon reduction limit. They first designed the DNA on a computer and then chemically synthesized and assembled 100 kb fragments in the S. cerevisiae vector, which were then taken up by E. coli and integrated directly into the corresponding natural regions of the genome (see Figure 1). By iterating this process 5 times, the 500 kb DNA segment can be completely replaced by synthetic DNA. Using this method, the team generated eight E. coli strains, each carrying a synthetic DNA segment covering different genomic regions. The researchers then piece together these segments by "joining" to synthesize the complete genome.
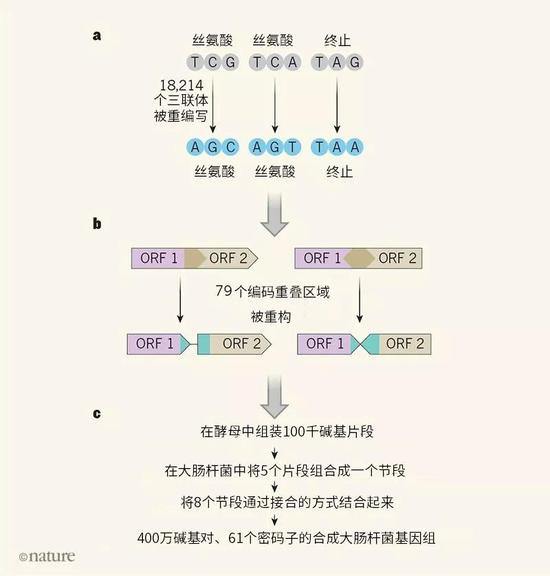
Figure 1 | Design and construction of a recoded genome. a. Fredens et al. reprogrammed the 3 base triplet (codon) of the E. coli genome, replacing the TCG and TCA encoding serine and the stop codon TAG representing the termination of the protein coding sequence into a password with the same function. Sub AGC, AGT and TAA.
b. At some sites in the genome, open reading frames (ORFs, ie, protein coding regions) overlap, and changes to an ORF codon may result in unexpected changes in overlapping regions. Fredens et al. "reconstruct" these ORFs to separate them, as shown by ORF1 and ORF2 (the "read" direction of the two ORFs on the left is the same, and the two reads on the right are opposite).
c. The redesigned DNA can be assembled into 100 kilobase fragments in Saccharomyces cerevisiae; these fragments are then combined into segments and integrated into the genome of E. coli. Combine these segments to obtain a complete functional synthetic genome.
This large-scale construction project has achieved amazing success, and the off-target mutation rate is low, but there are also some challenges. Many of the genes in the E. coli genome partially overlap with other genes, and the researchers found that 91 overlapping regions contain codons that need to be altered. This is complicated because synonymous substitutions in a protein coding sequence may alter the amino acids encoded by the overlapping sequences. To solve this problem, the research team “reconstructed†79 loci in the genome. By copying this sequence, they separated the overlapping coding sequences into a single recoding sequence (see Figure 1). Although this method is basically successful, there are some places that require very careful error checking because refactoring may alter gene regulation.
The resulting strain was confirmed to be viable and could grow in a variety of typical laboratory settings, but was slightly slower than the native strain. The new strain no longer requires the stop codon TAG and the serine codons TCG and TCA, so the cellular machinery that recognizes these codons can also be removed or reused to recruit "non-standard" amino acids - the majority of living cells needed Amino acids other than 20 common amino acids. The researchers used E. coli with only 63 codons to confirm the usefulness of recruiting non-standard amino acids. For biotechnology projects, non-standard amino acids can be written into the desired sequence positions, providing residues that can participate in chemical reactions in which natural proteins cannot participate. Moreover, since the genetic code supporting the operation of synthetic E. coli cells is slightly different from other cells in the natural world, the readable DNA-encoded information of the input and output of synthetic E. coli will become limited, and thus bring about biosafety. benefit. All of the above applications will be further extended in the new 61 codons of E. coli, which has the potential to encode more than one non-standard amino acid and build a more stringent genetic firewall (because of 3 of the 64 codons) Codons are no longer recognized).
The synthesis of a 4 million base pair genome and the reduction of the genetic code to 61 codons have created the latest record in synthetic genomics, but this record may not last long. The International Sc2.0 Alliance is trying to synthesize all 16 chromosomes of the 12 million base pair Saccharomyces cerevisiae genome, the first synthetic genome of eukaryotes including plants, animals and fungi. At the same time, the Alliance is also experimenting with the synthesis of an E. coli genome with only 57 codons and a synthetic genome with two fewer codons than the native Salmonella Typhimurium. If successful, bacteria with synthetic genomes in the future are expected to be used as cell-based technologies in the human gut.
From a purely technical point of view, the most noteworthy point of these different studies is that they have very similar processes for constructing synthetic genomes, in which the kilobase segments of synthetic DNA (by homologous recombination) are assembled in yeast cells. The 50-100 kilobase fragment is used to replace the native sequence in the target organism with these fragments (by selection of recombinant methods). The standardization of the method is conducive to the automation of some of the steps, allowing more teams to join the research. Genomic minimization and codon reduction are only preliminary applications of this technology, and in the future, this technology may generate functionally recombined genomes, or "customized" genomes that can direct cells to perform specific tasks.
Other Medical Consumables,Medical Equipment,Medical Equipment Consumables,Medical Consumable Products
Shandong Zhushi Pharmaceutical Group Co.,LTD , https://www.sdzs-medical.com